BPDL results on Synthetic datasets - binary images¶
Presenting results from starting version of BPDL
[1]:
%matplotlib inline
import os, sys, glob
import pandas, numpy
from skimage import io
import matplotlib.pylab as plt
from matplotlib import gridspec
sys.path += [os.path.abspath('.'), os.path.abspath('..')] # Add path to root
import bpdl.utilities as utils
import bpdl.data_utils as tl_data
/usr/local/lib/python2.7/dist-packages/matplotlib/__init__.py:1405: UserWarning:
This call to matplotlib.use() has no effect because the backend has already
been chosen; matplotlib.use() must be called *before* pylab, matplotlib.pyplot,
or matplotlib.backends is imported for the first time.
warnings.warn(_use_error_msg)
[2]:
p_results = utils.update_path('results')
p_csv = os.path.join(p_results, 'experiments_synth_APDL_binary_overall.csv')
print (os.path.exists(p_csv), '<-', p_csv)
p_data = '/mnt/F464B42264B3E590/TEMP'
DATASET = 'atomicPatternDictionary_v0'
print (os.path.exists(p_data), '<-', p_data)
True <- results/experiments_synth_APDL_binary_overall.csv
True <- /mnt/F464B42264B3E590/TEMP
Loading data¶
[3]:
df_all = pandas.read_csv(p_csv, index_col=None)
df_all.sort(['nb_labels'], inplace=True)
print ('-> loaded DF with', len(df_all), 'items and columns:\n', df_all.columns.tolist())
d_unique = {col: df_all[col].unique().tolist()
for col in ['dataset', 'gc_regul', 'gc_reinit', 'init_tp', 'ptn_split']}
print ('-> unique:', d_unique)
-> loaded DF with 11683 items and columns:
['atlas_ARS', 'case', 'class', 'computer', 'dataset', 'folders', 'gc_regul', 'gc_reinit', 'init_tp', 'max_iter', 'name', 'nb_workers', 'nb_labels', 'nb_res', 'nb_runs', 'nb_samples', 'overlap_major', 'path_exp', 'path_in', 'path_out', 'ptn_compact', 'ptn_split', 'reconstruct_diff', 'subfiles', 'time', 'tol', 'type']
-> unique: {'init_tp': ['msc2', 'GTd', 'msc', 'rnd', 'msc1', 'GT', 'OWSr', 'OWS', 'GWS'], 'ptn_split': [False, True], 'gc_reinit': [True, nan], 'gc_regul': [0.001, 0.0, 9.999999999999999e-16, 1e-09], 'dataset': ['datasetBinary_noise', 'datasetBinary_raw', 'datasetBinary_defNoise', 'datasetBinary_deform']}
/usr/local/lib/python2.7/dist-packages/IPython/kernel/__main__.py:2: FutureWarning: sort(columns=....) is deprecated, use sort_values(by=.....)
from IPython.kernel.zmq import kernelapp as app
[5]:
atlas = tl_data.dataset_compose_atlas(os.path.join(p_data, DATASET))
plt.imshow(atlas, interpolation='nearest', cmap=plt.cm.jet)
_ = plt.title('Atlas; unique {}'.format(numpy.unique(atlas).shape))

[6]:
print('path_in:', df_all['path_in'].unique())
print('gc_regul:', df_all['gc_regul'].unique())
path_in: [ '/datagrid/Medical/microscopy/drosophila/synthetic_data/atomicPatternDictionary_v0'
'/datagrid/Medical/microscopy/drosophila/synthetic_data/atomicPatternDictionary_v1'
'/datagrid/Medical/microscopy/drosophila/synthetic_data/atomicPatternDictionary_v2'
'/datagrid/Medical/microscopy/drosophila/synthetic_data/atomicPatternDictionary_v3']
gc_regul: [ 1.00000000e-03 0.00000000e+00 1.00000000e-15 1.00000000e-09]
Dependency on number of used patterns¶
take out the series with various param combination
[7]:
GC_REGUL = 0.0
df_select = df_all[df_all['path_in'].str.endswith(DATASET)]
df_select = df_select[df_select['gc_regul'] == GC_REGUL]
# df_select = df_select[df_select['ptn_split'] == False]
print ('selected records:', len(df_select))
df_res = pandas.DataFrame()
for v, df_gr0 in df_select.groupby('dataset'):
for v1, df_gr1 in df_gr0.groupby('init_tp'):
for v2, df_gr2 in df_gr1.groupby('ptn_split'):
# for v1, df_gr1 in df_gr0.groupby('overlap_major'):
name = '"{}", split ({})'.format(v1, int(v2))
d = {'dataset': v, 'name': name}
cols = ['nb_labels', 'atlas_ARS', 'reconstruct_diff', 'time', 'subfiles']
d.update({col: df_gr2[col].tolist() for col in cols})
df_res = df_res.append(d, ignore_index=True)
df_res = df_res.set_index('name')
# df_res.to_csv(os.path.join(os.path.dirname(p_csv), 'synth_APDL_%s_gc_%f.csv' % (DATASET, GC_REGUL)))
print ('number of rows:', len(df_res), 'columns:', df_res.columns.tolist())
selected records: 1152
number of rows: 72 columns: ['atlas_ARS', 'dataset', 'nb_labels', 'reconstruct_diff', 'subfiles', 'time']
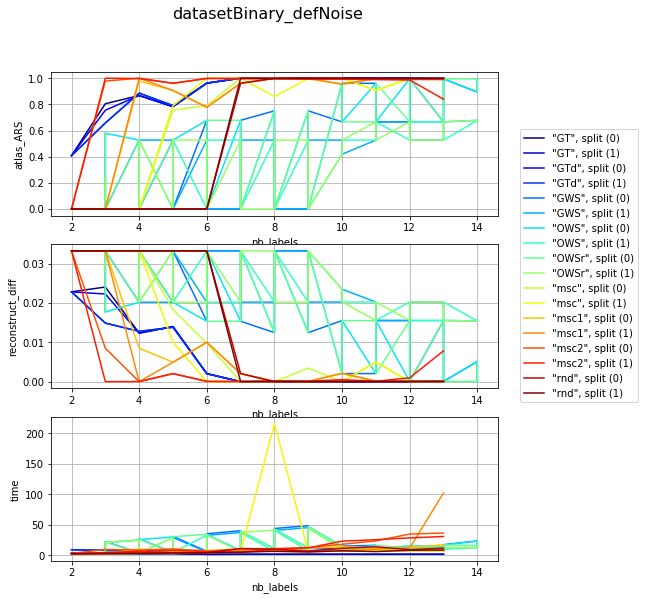
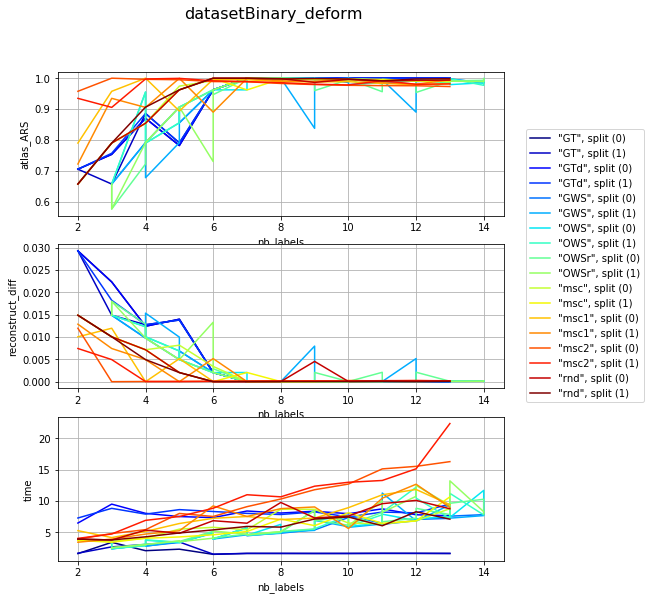
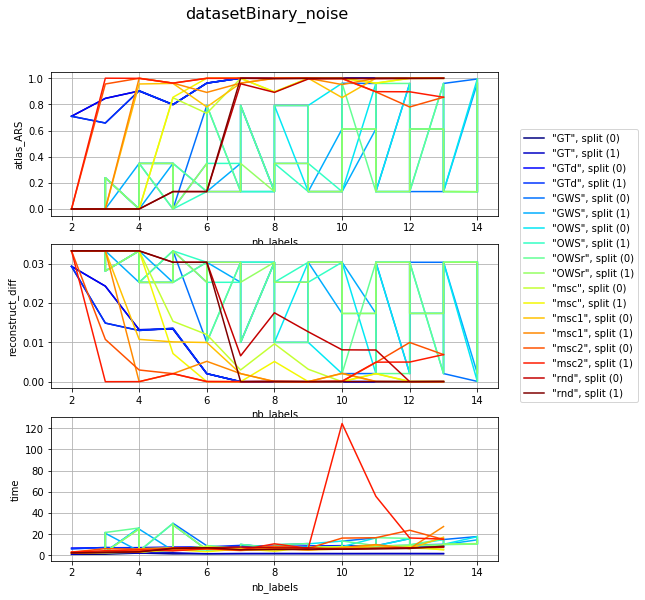
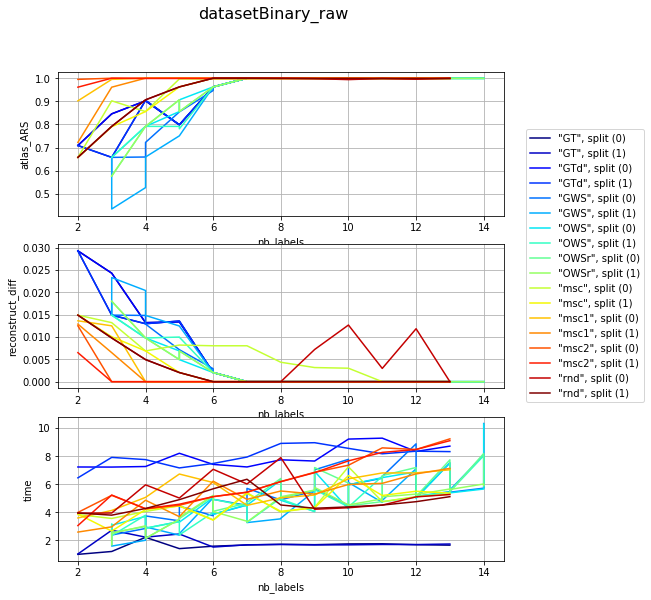
visualization per dataset (difficulty) and different param combination; sub fules corelate with mean number of iterartions ~ (subfiles / 2)
[13]:
# ADD graph ['nb_lbs', 'atlas_ARS', 'reconstruct_diff', 'subfiles'] and name as curve
l_graphs = ['atlas_ARS', 'reconstruct_diff', 'time']
for v, df_group in df_res.groupby('dataset'):
clrs = plt.cm.jet(numpy.linspace(0, 1, len(df_group)))
fig, axarr = plt.subplots(len(l_graphs), 1, figsize=(8, len(l_graphs) * 3))
fig.suptitle('{}'.format(v), fontsize=16)
for i, col in enumerate(l_graphs):
for j, (idx, row) in enumerate(df_group.iterrows()):
axarr[i].plot(row['nb_labels'], row[col], label=idx, color=clrs[j])
axarr[i].set_xlabel('nb_labels'), axarr[i].set_ylabel(col), axarr[i].grid()
plt.legend(bbox_to_anchor=(1.05, len(l_graphs)), loc=2, borderaxespad=0.)
[ ]: